How to Write Again to Csv File python
Still Saving Your Data in CSV? Try these other options
Learn how to save your data in different formats (CSV, compressions, Pickle, and Parquet) to save storage and reduce read/write time, and money
One of the most popular file formats for saving your dataframe is CSV (Comma-Separated Values). CSV is well known and widely supported on various platforms. However, while CSV is ubiquitous, it is not a suitable format when you are dealing with large amount of data — the size will get prohibitively large, and extracting data from it becomes painfully slow.
In this article, I will walk you through some of the options you have when saving your dataframe to storage. In particular, I will discuss saving your dataframe as:
- CSV file
- CSV file with compression applied
- Binary file using the pickle module
- Binary file using the pickle module, and with compression applied
- HDF file
- Parquet
Our Sample Dataset
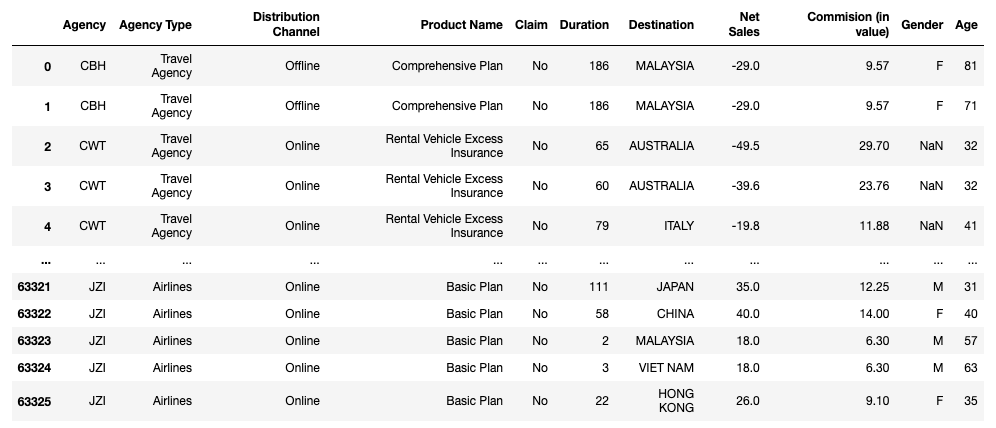
My sample dataset for this article is named travel insurance.csv (source: https://www.kaggle.com/mhdzahier/travel-insurance). It has 63,326 rows and 11 columns, with a mixture of object, int64, and float64 columns. Let's load it up using Pandas as a DataFrame:
import pandas as pd
import os filename = 'travel insurance'
df = pd.read_csv(filename + '.csv') display(df)
display(df.shape) # (63326, 11)
display(df.dtypes)
# Agency object
# Agency Type object
# Distribution Channel object
# Product Name object
# Claim object
# Duration int64
# Destination object
# Net Sales float64
# Commision (in value) float64
# Gender object
# Age int64
# dtype: object
To record the file size, write and read time for each data format, I will create a dataframe with four columns:
df_results = pd.DataFrame(columns=
['method', 'file_size', 'write_time', 'read_time'])
display(df_results) 
I will also create a helper function to append a row to the dataframe containing the details of each experiment:
def add_result(df_results, method, file_size,
write_time, read_time):
row = {
'method': method,
'file_size': file_size,
'write_time': write_time,
'read_time': read_time
}
return df_results.append(pd.Series(row), ignore_index = True) Saving as CSV Files
The first method that I want to try is save the dataframe back as a CSV file and then read it back. The following block of code does the following:
- Save the dataframe as a CSV file
- Get the size of the physical CSV file
- Load the CSV file again as a dataframe
- Write the method used, size of the file, average time taken to write the dataframe to file and load it back again, to the
df_resultsdataframe
#---saving---
result_save = %timeit -n5 -r5 -o df.to_csv(filename + '_csv.csv') #---get the size of file---
filesize = os.path.getsize(filename + '_csv.csv') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_csv(filename + '_csv.csv') #---save the result to the dataframe---
df_results = add_result(df_results,
'CSV',
filesize,
result_save.average,
result_read.average)
df_results
In particular, I used the %timeit magic command to record the time that the system spent executing a statement. For example, the following line records how much time the system spent saving the dataframe to a CSV file:
%timeit -n5 -r5 -o df.to_csv(filename + '_csv.csv') The %timeit magic command has the following options:
- The
-n5option means you want to run the statement 5 times in a loop. - The
-r5means run the loop 5 times and take the best result. This allows us to obtain a mean operation time for writing and reading the file. - The -o option indicates that you want the timing result to be returned instead of printing it out. In this case, the result is passed to the
result_savevariable for the write part andresult_readfor the read part. - With the result returned from the
%timeitcommand, you can use theaverageproperty to obtain the average time needed to perform the write and read operations.
With the results obtained for the writing and reading, you can now call the add_result() function to add the result to the dataframe. For this part, I obtained the following result:
165 ms ± 7.11 ms per loop (mean ± std. dev. of 5 runs, 5 loops each)
50.4 ms ± 9.32 ms per loop (mean ± std. dev. of 5 runs, 5 loops each) The df_result dataframe looks like this:

The file size is in megabytes (MB) and the times are in seconds.
Saving CSV without the Index
By default, the index of a dataframe is saved when persisting it as a CSV file. However, most of the time it is not necessary to save the index (which is just a series of running numbers) to the CSV file. And so we shall omit it this time round:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_csv(filename + '_csv.csv', \
index=False) #---get the size of file---
filesize = os.path.getsize(filename + '_csv.csv') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_csv(filename + '_csv.csv') #---save the result to the dataframe---
df_results = add_result(df_results,
'CSV No Index',
filesize,
result_save.average,
result_read.average) df_results
Here's the result:
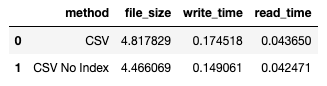
You can see that there is a slight reduction in file size, and the write and read times are both shortened.
Using Compression with CSV
Pandas supports compression when you save your dataframes to CSV files. Specifically, Pandas supports the following compression algorithms:
- gzip
- bz2
- zip
- xz
Let's see how compression will help in the file size as well as the write and read times.
GZIP
To use compression when you save your dataframes as CSV, set the compression parameter to the algorithm that you want to use. Likewise, when you load a compressed CSV files, set the compression parameter to the algorithm that was used to compress the file:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_csv(filename + '.gzip', \
index=False, \
compression='gzip') #---get the size of file---
filesize = os.path.getsize(filename + '.gzip') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_csv(filename + '.gzip', \
compression='gzip') #---save the result to the dataframe---
df_results = add_result(df_results,
'CSV No Index (GZIP)',
filesize,
result_save.average,
result_read.average) df_results
Here's the result:
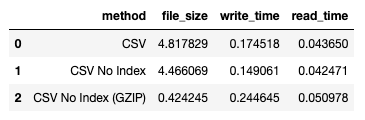
Quite a drastic reduction in file size. The write time is a bit longer, but the read time is comparable to the unzipped version.
For more information on GZIP, refer to: https://en.wikipedia.org/wiki/Gzip
BZ2
Another popular file compression algorithm is known as BZ2. The following code snippet shows compressing the dataframe using BZ2:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_csv(filename + '.bz2', \
index=False, \
compression='bz2') #---get the size of file---
filesize = os.path.getsize(filename + '.bz2') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_csv(filename + '.bz2', \
compression='bz2') #---save the result to the dataframe---
df_results = add_result(df_results,
'CSV No Index (BZ2)',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:
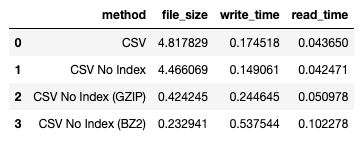
You can observe that BZ2 results in a better compression ratio compared to GZIP, but the corresponding write and read times are slower.
For more information on BZ2, refer to: https://en.wikipedia.org/wiki/Bzip2
Pickle
CSV files are plain-text files (except when they are compressed), and this makes them ubiquitous, and supported on all platforms and all major software. However, the main drawback of CSV is its size. Instead of saving your dataframe as a plain text file, you can save your dataframe as a binary file.
In Python, you can use the pickle module to persist your data (including your dataframe) as a binary file. The pickle module serializes your objects in Python into a binary file and deserializes the binary file back into an object in Python.
Let's try saving our dataframe using pickle:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_pickle(filename + '.pkl') #---get the size of file---
filesize = os.path.getsize(filename + '.pkl') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_pickle(filename + '.pkl') #---save the result to the dataframe---
df_results = add_result(df_results,
'Pickle',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:

The file size is smaller than the CSV file, but larger than if you were to use compression on the CSV files. However, notice that write and read times are the fastest up to this point.
Using Pickle with compression
Like saving as CSV, you can also use compression with pickle. First, let's use GZIP:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_pickle(filename + '.pkl', \
compression='gzip') #---get the size of file---
filesize = os.path.getsize(filename + '.pkl') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_pickle(filename + '.pkl', \
compression='gzip') #---save the result to the dataframe---
df_results = add_result(df_results,
'Pickle (GZIP)',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:

Then, we use BZ2:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_pickle(filename + '.pkl', \
compression='bz2') #---get the size of file---
filesize = os.path.getsize(filename + '.pkl') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_pickle(filename + '.pkl', \
compression='bz2') #---save the result to the dataframe---
df_results = add_result(df_results,
'Pickle (BZ2)',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:
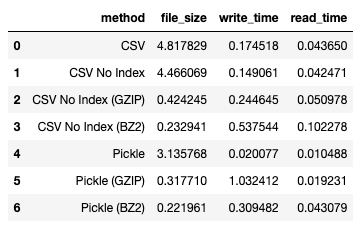
Next, we use zip:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_pickle(filename + '.pkl', \
compression='zip') #---get the size of file---
filesize = os.path.getsize(filename + '.pkl') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_pickle(filename + '.pkl', \
compression='zip') #---save the result to the dataframe---
df_results = add_result(df_results,
'Pickle (ZIP)',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:
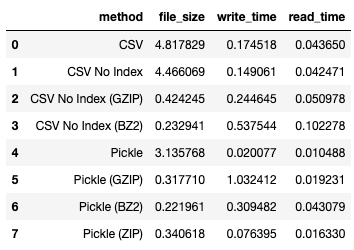
For more information on zip, refer to: https://en.wikipedia.org/wiki/ZIP_(file_format)
Finally, we use xz:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_pickle(filename + '.pkl', \
compression='xz') #---get the size of file---
filesize = os.path.getsize(filename + '.pkl') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_pickle(filename + '.pkl', \
compression='xz') #---save the result to the dataframe---
df_results = add_result(df_results,
'Pickle (xz)',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:
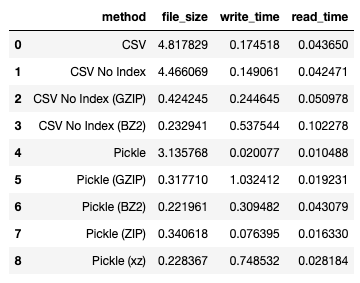
For more information on xz, refer to: https://en.wikipedia.org/wiki/XZ_Utils
As you can see from the results, using compressions with pickle greatly reduces the size of the file, while the write and read times increase a little bit compared to without using compression.
HDF
Another file format that you can use to save your dataframe is HDF — Hierarchical Data Format. HDF is an open source file format that supports large, complex, heterogeneous data. HDF uses a structure similar to a file directory to organize your data within the file.
Think of HDF as a file structure that allows you to store different dataframes, all in a single physical file.
The following code snippet saves the dataframe as a HDF file:
#---saving---
result_save = %timeit -n5 -r5 -o df.to_hdf(filename + '.h5', \
key='key', \
mode='w') #---get the size of file---
filesize = os.path.getsize(filename + '.h5') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_hdf(filename + '.h5', \
key='key', \
mode='r') #---save the result to the dataframe---
df_results = add_result(df_results,
'HDF',
filesize,
result_save.average,
result_read.average) df_results
I obtained the following result:

As you can observe, saving as HDF does not really reduce the file size, even though the write time is better than raw CSV.
Parquet
The last file format that I want to discuss is Parquet. So what is Parquet, or more accurately, Apache Parquet ?
Apache Parquet is a file format that is designed to support fast data processing for complex data. It is an open source file format under the Apache Hadoop license, and is compatible with most Hadoop processing frameworks. Parquet is self-describing — metadata including the schema and structure is embedded within each file.
More importantly, Parquet stores your data in columns, rather than rows.
How Parquet stores your data
Consider the following dataframe with three columns:
When you save the dataframe as a CSV file, it uses row-based storage. When a CSV file is loaded into a dataframe, each row is loaded one at a time, and each row contains three different data types:
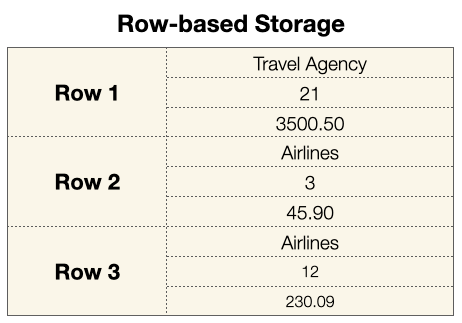
On the other hand, Parquet stores your data using column-based storage. Each column of data is organized as a column, of a specific data type:
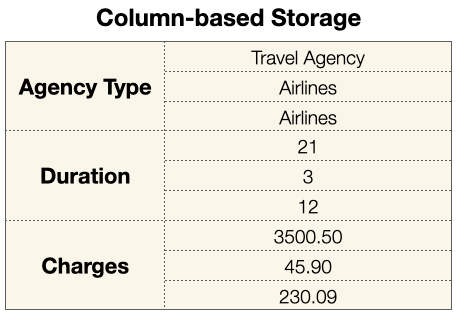
In short, when you organize your data in column-based storage, your file would be more lightweight since all similar data types are grouped together, and you can apply compressions to each column. More importantly, using column-based storage makes its really efficient to extract specific columns, something that you do very often for data analytics projects.
To use parquet you need to install the fastparquet module:
!pip install fastparquet The following code snippet saves the dataframe into a parquet file and then load it back up:
# !parq Building_Permits.parquet --head 10 #---saving---
result_save = %timeit -n5 -r5 -o df.to_parquet(filename + \
'.parquet', \
engine='fastparquet') #---get the size of file---
filesize = os.path.getsize(filename + '.parquet') / 1024**2 #---load---
result_read = %timeit -n5 -r5 -o pd.read_parquet(filename + \
'.parquet') #---save the result to the dataframe---
df_results = add_result(df_results,
'Parquet',
filesize,
result_save.average,
result_read.average) df_results
I get the following results:
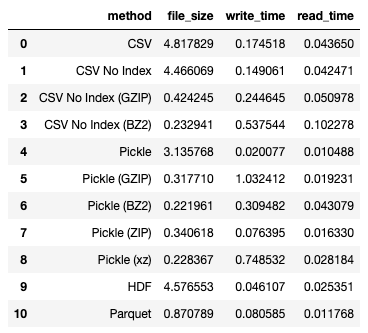
From the result, you can see the the file size is drastically reduced (though not as much as using compression on CSV or pickle, but the reduction is still significant), and the write and read time is one of the fastest.
So why is Parquet useful and why should you use it? There are a few compelling reasons why you should store your data using Parquet rather than CSVs:
- Cloud services (such as AWS and Google) charge based on the size of your data. Parquet files are much smaller than CSV files and this means you pay lesser for cloud storage.
- Cloud services also charge based on data scanned per query. In the above results, you can see that write and read time are faster than CSV files. Lower scanning time means lower cloud charges.
To see how Parquet performs in comparison with CSV files when loading specific columns, let's try the following experiment where we will load up two columns (Agency Type and Product Name) from three CSV files (one uncompressed, one compressed with BZ2 and one compressed with GZIP) and one parquet file:
%timeit -n5 -r5 -o pd.read_csv(filename + '_csv.csv', \
usecols=['Agency Type','Product Name']) %timeit -n5 -r5 -o pd.read_csv(filename + '.bz2', \
usecols=['Agency Type','Product Name'], \
compression='bz2') %timeit -n5 -r5 -o pd.read_csv(filename + '.gzip', \
usecols=['Agency Type','Product Name'], \
compression='gzip') %timeit -n5 -r5 -o pd.read_parquet(filename + '.parquet', \
columns=['Agency Type','Product Name'])
The above statements returns the following results:
27.4 ms ± 6.23 ms per loop (mean ± std. dev. of 5 runs, 5 loops each) 77.4 ms ± 3.26 ms per loop (mean ± std. dev. of 5 runs, 5 loops each) 35 ms ± 2.15 ms per loop (mean ± std. dev. of 5 runs, 5 loops each) 4.72 ms ± 1.73 ms per loop (mean ± std. dev. of 5 runs, 5 loops each)
The last result (using Parquet) beats the rest hands down. Using Parquet takes on average 4.72 ms to retrieve the two columns, compared to about 27 to 77 ms for CSV files.
Finding the Best Method for Each Task
Now that we have the statistics for the various methods, we can very easily find out which is the method that is most ideal for each task.
If you want to minimize read time, saving using pickle is the best solution:
df_results.sort_values(by='read_time') 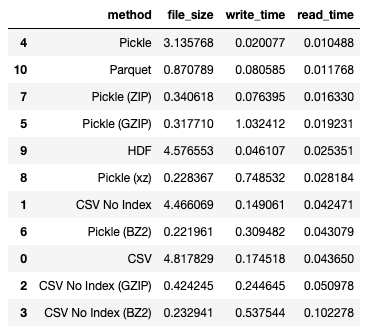
Observe that Parquet is a close second in terms of read time. Moreover, it also greatly reduces the file size. As mentioned in the previous section, minimizing read time and storage can be critical to reduce your costs when you store your data on cloud services.
If you want to minimize write time, seems like using pickle is also the best way to save your dataframe:
df_results.sort_values(by='write_time') 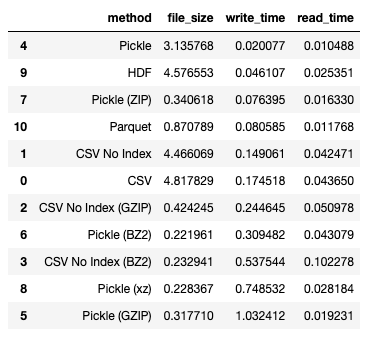
If you want to minimize file size of your dataframe, use pickle together with the bz2 compression algorithm:
df_results.sort_values(by='file_size') 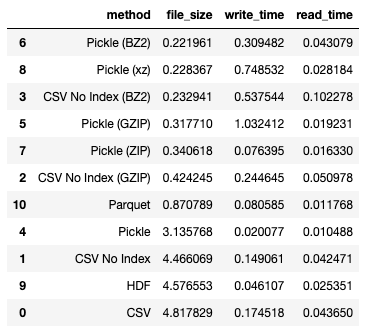
Summary
This article discusses some of the file formats that you can use to save your dataframes. This is by no means an exhaustive test, but it gives you an overall feel of how each file format performs. It is important to note that the results I have obtained in this article is specific to my dataset. Different dataset may have different results, depending on the type of data you have. If your dataset have a lot of object (text) columns, compression will work really well. What is important is — test your dataset using the various methods described in this article and draw your own conclusions. In particular, compare the statistics of your most frequently performed operations, and decide which methods suits your data better.
grahampravall1951.blogspot.com
Source: https://towardsdatascience.com/still-saving-your-data-in-csv-try-these-other-options-9abe8b83db3a
Postar um comentário for "How to Write Again to Csv File python"